Visualizations#
Tensorboard visuals#
- monai.visualize.img2tensorboard.add_animated_gif(writer, tag, image_tensor, max_out=3, frame_dim=-3, scale_factor=1.0, global_step=None)[source]#
Creates an animated gif out of an image tensor in ‘CHWD’ format and writes it with SummaryWriter.
- Parameters:
writer – Tensorboard SummaryWriter to write to
tag – Data identifier
image_tensor – tensor for the image to add, expected to be in CHWD format
max_out – maximum number of image channels to animate through
frame_dim – the dimension used as frames for GIF image, expect input data shape as CHWD, default to -3 (the first spatial dim)
scale_factor – amount to multiply values by. If the image data is between 0 and 1, using 255 for this value will scale it to displayable range
global_step – Global step value to record
- monai.visualize.img2tensorboard.make_animated_gif_summary(tag, image, writer=None, max_out=3, frame_dim=-3, scale_factor=1.0)[source]#
Creates an animated gif out of an image tensor in ‘CHWD’ format and returns Summary.
- Parameters:
tag – Data identifier
image – The image, expected to be in CHWD format
writer – the tensorboard writer to plot image
max_out – maximum number of image channels to animate through
frame_dim – the dimension used as frames for GIF image, expect input data shape as CHWD, default to -3 (the first spatial dim)
scale_factor – amount to multiply values by. if the image data is between 0 and 1, using 255 for this value will scale it to displayable range
- monai.visualize.img2tensorboard.plot_2d_or_3d_image(data, step, writer, index=0, max_channels=1, frame_dim=-3, max_frames=24, tag='output')[source]#
Plot 2D or 3D image on the TensorBoard, 3D image will be converted to GIF image.
Note
Plot 3D or 2D image(with more than 3 channels) as separate images. And if writer is from TensorBoardX, data has 3 channels and max_channels=3, will plot as RGB video.
- Parameters:
data – target data to be plotted as image on the TensorBoard. The data is expected to have ‘NCHW[D]’ dimensions or a list of data with CHW[D] dimensions, and only plot the first in the batch.
step – current step to plot in a chart.
writer – specify TensorBoard or TensorBoardX SummaryWriter to plot the image.
index – plot which element in the input data batch, default is the first element.
max_channels – number of channels to plot.
frame_dim – if plotting 3D image as GIF, specify the dimension used as frames, expect input data shape as NCHWD, default to -3 (the first spatial dim)
max_frames – if plot 3D RGB image as video in TensorBoardX, set the FPS to max_frames.
tag – tag of the plotted image on TensorBoard.
Class activation map#
- class monai.visualize.class_activation_maps.CAM(nn_module, target_layers, fc_layers='fc', upsampler=<function default_upsampler>, postprocessing=<function default_normalizer>)[source]#
Compute class activation map from the last fully-connected layers before the spatial pooling. This implementation is based on:
Zhou et al., Learning Deep Features for Discriminative Localization. CVPR ‘16, https://arxiv.org/abs/1512.04150
Examples
import torch # densenet 2d from monai.networks.nets import DenseNet121 from monai.visualize import CAM model_2d = DenseNet121(spatial_dims=2, in_channels=1, out_channels=3) cam = CAM(nn_module=model_2d, target_layers="class_layers.relu", fc_layers="class_layers.out") result = cam(x=torch.rand((1, 1, 48, 64))) # resnet 2d from monai.networks.nets import seresnet50 from monai.visualize import CAM model_2d = seresnet50(spatial_dims=2, in_channels=3, num_classes=4) cam = CAM(nn_module=model_2d, target_layers="layer4", fc_layers="last_linear") result = cam(x=torch.rand((2, 3, 48, 64)))
N.B.: To help select the target layer, it may be useful to list all layers:
for name, _ in model.named_modules(): print(name)
- __init__(nn_module, target_layers, fc_layers='fc', upsampler=<function default_upsampler>, postprocessing=<function default_normalizer>)[source]#
- Parameters:
nn_module – the model to be visualized
target_layers – name of the model layer to generate the feature map.
fc_layers – a string or a callable used to get fully-connected weights to compute activation map from the target_layers (without pooling). and evaluate it at every spatial location.
upsampler – An upsampling method to upsample the output image. Default is N dimensional linear (bilinear, trilinear, etc.) depending on num spatial dimensions of input.
postprocessing – a callable that applies on the upsampled output image. Default is normalizing between min=1 and max=0 (i.e., largest input will become 0 and smallest input will become 1).
- compute_map(x, class_idx=None, layer_idx=-1, **kwargs)[source]#
Compute the actual feature map with input tensor x.
- Parameters:
x – input to nn_module.
class_idx – index of the class to be visualized. Default to None (computing class_idx from argmax)
layer_idx – index of the target layer if there are multiple target layers. Defaults to -1.
- Returns:
activation maps (raw outputs without upsampling/post-processing.)
- class monai.visualize.class_activation_maps.GradCAM(nn_module, target_layers, upsampler=<function default_upsampler>, postprocessing=<function default_normalizer>, register_backward=True)[source]#
Computes Gradient-weighted Class Activation Mapping (Grad-CAM). This implementation is based on:
Selvaraju et al., Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization, https://arxiv.org/abs/1610.02391
Examples
import torch # densenet 2d from monai.networks.nets import DenseNet121 from monai.visualize import GradCAM model_2d = DenseNet121(spatial_dims=2, in_channels=1, out_channels=3) cam = GradCAM(nn_module=model_2d, target_layers="class_layers.relu") result = cam(x=torch.rand((1, 1, 48, 64))) # resnet 2d from monai.networks.nets import seresnet50 from monai.visualize import GradCAM model_2d = seresnet50(spatial_dims=2, in_channels=3, num_classes=4) cam = GradCAM(nn_module=model_2d, target_layers="layer4") result = cam(x=torch.rand((2, 3, 48, 64)))
N.B.: To help select the target layer, it may be useful to list all layers:
for name, _ in model.named_modules(): print(name)
- compute_map(x, class_idx=None, retain_graph=False, layer_idx=-1, **kwargs)[source]#
Compute the actual feature map with input tensor x.
- Parameters:
x – input to nn_module.
class_idx – index of the class to be visualized. Default to None (computing class_idx from argmax)
layer_idx – index of the target layer if there are multiple target layers. Defaults to -1.
- Returns:
activation maps (raw outputs without upsampling/post-processing.)
- class monai.visualize.class_activation_maps.GradCAMpp(nn_module, target_layers, upsampler=<function default_upsampler>, postprocessing=<function default_normalizer>, register_backward=True)[source]#
Computes Gradient-weighted Class Activation Mapping (Grad-CAM++). This implementation is based on:
Chattopadhyay et al., Grad-CAM++: Improved Visual Explanations for Deep Convolutional Networks, https://arxiv.org/abs/1710.11063
- compute_map(x, class_idx=None, retain_graph=False, layer_idx=-1, **kwargs)[source]#
Compute the actual feature map with input tensor x.
- Parameters:
x – input to nn_module.
class_idx – index of the class to be visualized. Default to None (computing class_idx from argmax)
layer_idx – index of the target layer if there are multiple target layers. Defaults to -1.
- Returns:
activation maps (raw outputs without upsampling/post-processing.)
- class monai.visualize.class_activation_maps.ModelWithHooks(nn_module, target_layer_names, register_forward=False, register_backward=False)[source]#
A model wrapper to run model forward/backward steps and storing some intermediate feature/gradient information.
- __init__(nn_module, target_layer_names, register_forward=False, register_backward=False)[source]#
- Parameters:
nn_module – the model to be wrapped.
target_layer_names – the names of the layer to cache.
register_forward – whether to cache the forward pass output corresponding to target_layer_names.
register_backward – whether to cache the backward pass output corresponding to target_layer_names.
- monai.visualize.class_activation_maps.default_normalizer(x)[source]#
A linear intensity scaling by mapping the (min, max) to (1, 0). If the input data is PyTorch Tensor, the output data will be Tensor on the same device, otherwise, output data will be numpy array.
Note: This will flip magnitudes (i.e., smallest will become biggest and vice versa).
- Return type:
~NdarrayTensor
Occlusion sensitivity#
- class monai.visualize.occlusion_sensitivity.OcclusionSensitivity(nn_module, mask_size=16, n_batch=16, verbose=True, mode='gaussian', overlap=0.25, activate=True)[source]#
This class computes the occlusion sensitivity for a model’s prediction of a given image. By occlusion sensitivity, we mean how the probability of a given prediction changes as the occluded section of an image changes. This can be useful to understand why a network is making certain decisions.
As important parts of the image are occluded, the probability of classifying the image correctly will decrease. Hence, more negative values imply the corresponding occluded volume was more important in the decision process.
Two
torch.Tensorwill be returned by the__call__method: an occlusion map and an image of the most probable class. Both images will be cropped if a bounding box used, but voxel sizes will always match the input.The occlusion map shows the inference probabilities when the corresponding part of the image is occluded. Hence, more -ve values imply that region was important in the decision process. The map will have shape
BCHW(D)N, whereNis the number of classes to be inferred by the network. Hence, the occlusion for classican be seen withmap[...,i].The most probable class is an image of the probable class when the corresponding part of the image is occluded (equivalent to
occ_map.argmax(dim=-1)).See: R. R. Selvaraju et al. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. https://doi.org/10.1109/ICCV.2017.74.
Examples:
# densenet 2d from monai.networks.nets import DenseNet121 from monai.visualize import OcclusionSensitivity import torch model_2d = DenseNet121(spatial_dims=2, in_channels=1, out_channels=3) occ_sens = OcclusionSensitivity(nn_module=model_2d) occ_map, most_probable_class = occ_sens(x=torch.rand((1, 1, 48, 64)), b_box=[2, 40, 1, 62]) # densenet 3d from monai.networks.nets import DenseNet from monai.visualize import OcclusionSensitivity model_3d = DenseNet(spatial_dims=3, in_channels=1, out_channels=3, init_features=2, growth_rate=2, block_config=(6,)) occ_sens = OcclusionSensitivity(nn_module=model_3d, n_batch=10) occ_map, most_probable_class = occ_sens(torch.rand(1, 1, 6, 6, 6), b_box=[1, 3, -1, -1, -1, -1])
See also
monai.visualize.occlusion_sensitivity.OcclusionSensitivity.
- __init__(nn_module, mask_size=16, n_batch=16, verbose=True, mode='gaussian', overlap=0.25, activate=True)[source]#
Occlusion sensitivity constructor.
- Parameters:
nn_module – Classification model to use for inference
mask_size – Size of box to be occluded, centred on the central voxel. If a single number is given, this is used for all dimensions. If a sequence is given, this is used for each dimension individually.
n_batch – Number of images in a batch for inference.
verbose – Use progress bar (if
tqdmavailable).mode –
what should the occluded region be replaced with? If a float is given, that value will be used throughout the occlusion. Else,
gaussian,mean_imgandmean_patchcan be supplied:gaussian: occluded region is multiplied by 1 - gaussian kernel. In this fashion, the occlusion will be 0 at the center and will be unchanged towards the edges, varying smoothly between. When gaussian is used, a weighted average will be used to combine overlapping regions. This will be done using the gaussian (not 1-gaussian) as occluded regions count more.mean_patch: occluded region will be replaced with the mean of occluded region.mean_img: occluded region will be replaced with the mean of the whole image.
overlap – overlap between inferred regions. Should be in range 0<=x<1.
activate – if
True, do softmax activation if num_channels > 1 else dosigmoid. IfFalse, don’t do any activation. Ifcallable, use callable on inferred outputs.
- static constant_occlusion(x, val, mask_size)[source]#
Occlude with a constant occlusion. Multiplicative is zero, additive is constant value.
- Return type:
tuple[float,Tensor]
- static crop_meshgrid(grid, b_box, mask_size)[source]#
Crop the meshgrid so we only perform occlusion sensitivity on a subsection of the image.
- Return type:
tuple[MetaTensor,SpatialCrop,Sequence]
- static gaussian_occlusion(x, mask_size, sigma=0.25)[source]#
For Gaussian occlusion, Multiplicative is 1-Gaussian, additive is zero. Default sigma of 0.25 empirically shown to give reasonable kernel, see here: Project-MONAI/MONAI#5230.
- Return type:
tuple[Tensor,float]
- static predictor(cropped_grid, nn_module, x, mul, add, mask_size, occ_mode, activate, module_kwargs)[source]#
Predictor function to be passed to the sliding window inferer. Takes a cropped meshgrid, referring to the coordinates in the input image. We use the index of the top-left corner in combination
mask_sizeto figure out which region of the image is to be occluded. The occlusion is performed on the original image,x, usingcropped_region * mul + add.mulandaddare sometimes pre-computed (e.g., a constant Gaussian blur), or they are sometimes calculated on the fly (e.g., the mean of the occluded patch). For this reasonocc_modeis given. Lastly,activateis used to activate after each call of the model.- Parameters:
cropped_grid – subsection of the meshgrid, where each voxel refers to the coordinate of the input image. The meshgrid is created by the
OcclusionSensitivityclass, and the generation of the subset is determined bysliding_window_inference.nn_module – module to call on data.
x – the image that was originally passed into
OcclusionSensitivity.__call__.mul – occluded region will be multiplied by this. Can be
torch.Tensororfloat.add – after multiplication, this is added to the occluded region. Can be
torch.Tensororfloat.mask_size – Size of box to be occluded, centred on the central voxel. Should be a sequence, one value for each spatial dimension.
occ_mode – might be used to calculate
mulandaddon the fly.activate – if
True, do softmax activation if num_channels > 1 else dosigmoid. IfFalse, don’t do any activation. Ifcallable, use callable on inferred outputs.module_kwargs – kwargs to be passed onto module when inferring
Gradient-based saliency maps#
- class monai.visualize.gradient_based.GuidedBackpropGrad(model)[source]#
Based on Springenberg and Dosovitskiy et al. https://arxiv.org/abs/1412.6806, compute gradient-based saliency maps by backpropagating positive gradients and inputs (see
_AutoGradReLU).See also
Springenberg and Dosovitskiy et al. Striving for Simplicity: The All Convolutional Net (https://arxiv.org/abs/1412.6806)
- class monai.visualize.gradient_based.GuidedBackpropSmoothGrad(model, stdev_spread=0.15, n_samples=25, magnitude=True, verbose=True)[source]#
Compute gradient-based saliency maps based on both
GuidedBackpropGradandSmoothGrad.
- class monai.visualize.gradient_based.SmoothGrad(model, stdev_spread=0.15, n_samples=25, magnitude=True, verbose=True)[source]#
Compute averaged sensitivity map based on
n_samples(Gaussian additive) of noisy versions of the input imagex.See also
Smilkov et al. SmoothGrad: removing noise by adding noise https://arxiv.org/abs/1706.03825
- class monai.visualize.gradient_based.VanillaGrad(model)[source]#
Given an input image
x, calling this class will perform the forward pass, then set to zero all activations except one (defined byindex) and propagate back to the image to achieve a gradient-based saliency map.If
indexis None, argmax of the output logits will be used.See also
Simonyan et al. Deep Inside Convolutional Networks: Visualising Image Classification Models and Saliency Maps (https://arxiv.org/abs/1312.6034)
Utilities#
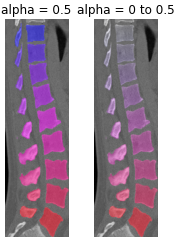
- monai.visualize.utils.blend_images(image, label, alpha=0.5, cmap='hsv', rescale_arrays=True, transparent_background=True)[source]#
Blend an image and a label. Both should have the shape CHW[D]. The image may have C==1 or 3 channels (greyscale or RGB). The label is expected to have C==1.
- Parameters:
image – the input image to blend with label data.
label – the input label to blend with image data.
alpha – this specifies the weighting given to the label, where 0 is completely transparent and 1 is completely opaque. This can be given as either a single value or an array/tensor that is the same size as the input image.
cmap – specify colormap in the matplotlib, default to hsv, for more details, please refer to: https://matplotlib.org/2.0.2/users/colormaps.html.
rescale_arrays – whether to rescale the array to [0, 1] first, default to True.
transparent_background – if true, any zeros in the label field will not be colored.
- monai.visualize.utils.matshow3d(volume, fig=None, title=None, figsize=(10, 10), frames_per_row=None, frame_dim=-3, channel_dim=None, vmin=None, vmax=None, every_n=1, interpolation='none', show=False, fill_value=nan, margin=1, dtype=<class 'numpy.float32'>, **kwargs)[source]#
Create a 3D volume figure as a grid of images.
- Parameters:
volume – 3D volume to display. data shape can be BCHWD, CHWD or HWD. Higher dimensional arrays will be reshaped into (-1, H, W, [C]), C depends on channel_dim arg. A list of channel-first (C, H[, W, D]) arrays can also be passed in, in which case they will be displayed as a padded and stacked volume.
fig – matplotlib figure or Axes to use. If None, a new figure will be created.
title – title of the figure.
figsize – size of the figure.
frames_per_row – number of frames to display in each row. If None, sqrt(firstdim) will be used.
frame_dim – for higher dimensional arrays, which dimension from (-1, -2, -3) is moved to the -3 dimension. dim and reshape to (-1, H, W) shape to construct frames, default to -3.
channel_dim – if not None, explicitly specify the channel dimension to be transposed to the last dimensionas shape (-1, H, W, C). this can be used to plot RGB color image. if None, the channel dimension will be flattened with frame_dim and batch_dim as shape (-1, H, W). note that it can only support 3D input image. default is None.
vmin – vmin for the matplotlib imshow.
vmax – vmax for the matplotlib imshow.
every_n – factor to subsample the frames so that only every n-th frame is displayed.
interpolation – interpolation to use for the matplotlib matshow.
show – if True, show the figure.
fill_value – value to use for the empty part of the grid.
margin – margin to use for the grid.
dtype – data type of the output stacked frames.
kwargs – additional keyword arguments to matplotlib matshow and imshow.
See also
Example
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> from monai.visualize import matshow3d # create a figure of a 3D volume >>> volume = np.random.rand(10, 10, 10) >>> fig = plt.figure() >>> matshow3d(volume, fig=fig, title="3D Volume") >>> plt.show() # create a figure of a list of channel-first 3D volumes >>> volumes = [np.random.rand(1, 10, 10, 10), np.random.rand(1, 10, 10, 10)] >>> fig = plt.figure() >>> matshow3d(volumes, fig=fig, title="List of Volumes") >>> plt.show()